How to warp¶
Running BigBrainWarp¶
First, install the package.
To run BigBrainWarp using docker, you should mount a working directory (using the -v argument). It is expected that your input data is housed in the mounted directory. The output will also be deposited in that directory.
# with docker (change the path to the local location of the mount accordingly)
docker run -it --rm -v /local/directory/with/data/:/BigBrainWarp/mount \
caseypaquola/bigbrainwarp bigbrainwarp
# without docker
bigbrainwarp
The following arguments can be used with BigBrainWarp
| Parameter | Description | Conditions | Options |
|---|---|---|---|
| in_space | Space of input data | Required | bigbrain, bigbrainsym, icbm, fsaverage, fs_LR |
| out_space | Space of output data | Required | bigbrain, bigbrainsym, icbm, fsaverage, fs_LR |
| wd | Path to working directory | Required | |
| desc | Descriptor that determines the naming of the output. See more details in Naming Conventions | Required. | |
| in_vol | Full path to input data, whole brain volume. | Requires either in_vol, or in_lh and in_rh | Permitted formats: mnc, nii or nii.gz |
| ih_lh | Full path to input data, left hemisphere surface. | Requires either in_vol, or in_lh and in_rh | Permitted formats: label.gii, annot, shape.gii, curv or txt |
| ih_rh | Full path to input data, left hemisphere surface. | Requires either in_vol, or in_lh and in_rh | Permitted formats: label.gii, annot, shape.gii, curv or txt |
| interp | Interpolation method | Required for in_vol. | Optional for txt input. Not permitted for other surface inputs. For in_vol, can be trilinear (default), tricubic, nearest or sinc. For txt, can be linear or nearest |
| out_type | Specifies whether output in surface or volume space | Optional function for bigbrain and bigbrainsym output. Otherwise, defaults to the same type as the input. | surface, volume |
| out_res | Resolution of output volume | Optional where out_type is volume. Default is 1. | Value provided in mm |
| out_dens | Density of output mesh | Optional where out_type is surface. Default is 164 | For fs_LR, 164 or 32 |
What’s under the hood?¶
The BigBrainWarp function currently wraps the following range of transformations. In C), the output template is not specified as it is assumed that either the input or the output is BigBrain.
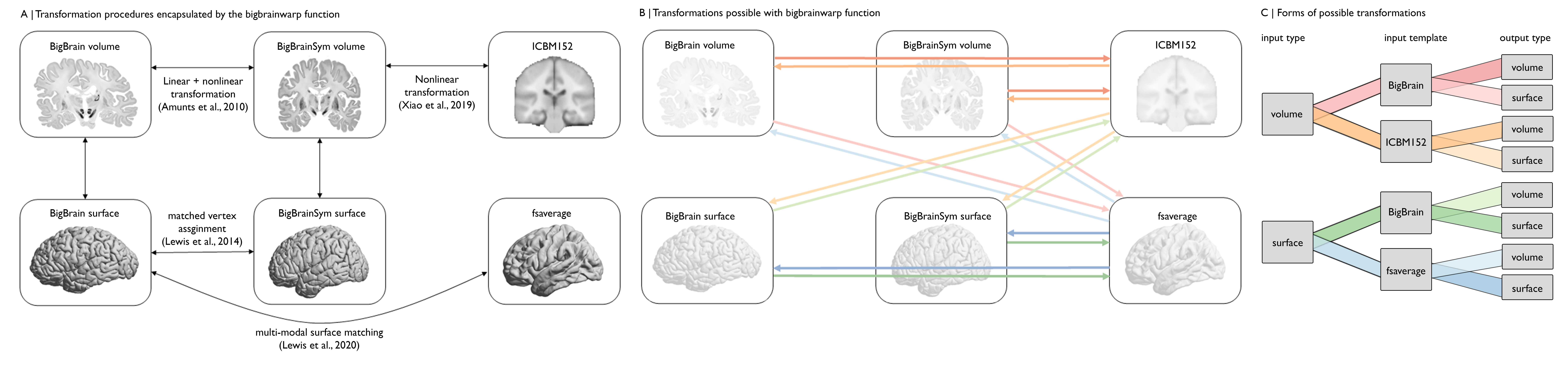
Each coloured path represents a distinct script called by bigbrainwarp. The method for each transformation is as follows:
| Input type | Input template | Output type | Colour | Method |
|---|---|---|---|---|
| Volume | BigBrain | Volume | Dark pink | Nonlinear volume-based transformation (Xiao et al.,) |
| Volume | BigBrain | Surface | Light pink | Volume-to-surface (HCP workbench) then multi-modal surface matching (Lewis et al., 2020) |
| Volume | ICBM152 | Volume | Dark orange | Nonlinear volume-based transformation (Xiao et al.,) |
| Volume | ICBM152 | Surface | Light orange | Nonlinear volume-based transformation (Xiao et al.,) then volume-to-surface (HCP workbench) |
| Surface | BigBrain | Volume | Dark green | Surface-to-volume (HCP workbench) then nonlinear volume-based transformation (Xiao et al.,) |
| Surface | BigBrain | Surface | Light green | Multi-modal surface matching (Lewis et al., 2020) |
| Surface | fsaverage | Volume | Dark blue | Multi-modal surface matching (Lewis et al., 2020) then surface-to-volume (HCP workbench) |
| Surface | fsaverage | Surface | Light blue | Multi-modal surface matching (Lewis et al., 2020) |
Example transformations in volume space¶
# for example, transformation of a bigbrain to icbm can take the form with a local installation
bigbrainwarp --in_vol data.nii --in_space bigbrain --interp trilinear --out_space icbm --out_res 0.5 --desc data --wd /project/
# or with the docker version - note how the working input volume is preprended with the "mount/" and the working directory is set to "mount"
docker run -it --rm -v /project/:/BigBrainWarp/mount caseypaquola/bigbrainwarp \
bigbrainwarp --in_vol mount/data.nii --in_space bigbrain --interp trilinear --out_space icbm --out_res 0.5 --desc data --wd mount
# in contrast, transformation from icbm to bigbrainsym could be
bigbrainwarp --in_vol data.mnc --in_space icbm --interp sinc --out_space bigbrainsym --desc data --wd /project/
BigBrainWarp utilises a recently published nonlinear transformation Xiao et al., (2019) If you use volume-based transformations in BigBrainWarp, please cite: Xiao, Y., et al. ‘An accurate registration of the BigBrain dataset with the MNI PD25 and ICBM152 atlases’. Sci Data 6, 210 (2019). https://doi.org/10.1038/s41597-019-0217-0
Example transformations for surface-based data¶
Surface-based transformation can be enacted using multi-modal surface matching; a spherical registration approach. Ongoing work by Lewis et al., involves optimisation of registration surafces between BigBrain and standard surface templates. These are available at ftp://bigbrain.loris.ca/BigBrainRelease.2015/BigBrainWarp_Support. More details on procedure can be also found on the following poster and slides. The functions currently support fsaverage and fs_LR as standard imaging templates for input or output.
# for example, transformation of a bigbrain to fsaverage can take the form
bigbrainwarp --in_lh lh.data.label.gii --in_rh rh.data.label.gii --in_space bigbrain --out_space fsaverage --desc data --wd /project/
# or with the docker version - note how the working inputs are preprended with the "mount/" and the working directory is set to "mount"
docker run -it --rm -v /project/:/BigBrainWarp/mount caseypaquola/bigbrainwarp \
bigbrainwarp --in_lh mount/lh.data.label.gii --in_rh mount/rh.data.label.gii --in_space bigbrain --out_space fsaverage --desc data --wd mount
# in contrast, transformation from icbm to bigbrainsym could be
bigbrainwarp --in_lh lh.data.label.txt --in_rh rh.data.label.txt --in_space fs_LR --interp linear --out_space bigbrain --out_dens 32 --desc data --wd /project/
If you use surface-based transformations in BigBrainWarp, please cite: Lewis, L.B., et al. ‘A multimodal surface matching (MSM) surface registration pipeline to bridge atlases across the MNI and the Freesurfer/Human Connectome Project Worlds’ OHBM, Virtual (2020)