Tutorial 1: Comparing BigBrain- and MRI-derived cortical gradients on a common surface¶
In this tutorial, we aim to inspect the convergence of large-scale cortical gradients of cytoarchitecture, microstructure and functional connectivity.
Full tutorial code ➡️ https://github.com/caseypaquola/BigBrainWarp/tree/master/tutorials/gradients
First, we need to identify the input data and the transformations necessary to examine BigBrain- and MRI-derived gradients on a common surface. For BigBrain, we can use microstructure profiles to resolve the cytoarchitectural gradients. This procedure will involve matrix manipulation that is infeasible with a 327684 x 327684 array, however, so we will need to reduce the number of microstructure profiles prior to computation of the cytoarchitectural gradients. For MRI, we can use microstructure profiles of quantitative T1 mapping and resting state fMRI timeseries from the MICs dataset. Individual subject data must be aligned to a standard surface, for example fsaverage, then again downsampled to a feasible number of parcels for cross-correlation and embedding. Finally, we will need to define an interpolation strategy from the downsampled BigBrain surface to the downsampled fsaverage surface.
Given the need to downsample BigBrain, we’ll start by performing mesh decimation on the BigBrain surface. Mesh decimation will decrease the number of vertices in the surface close to a prescribed number (in this case ~10,000) and retriangulate the surface, in such a way that preserves the overall shape. You can find the decimated mesh in BigBrainWarp/scripts/nn_surface_indexing.mat as the BB10 variable.
% mesh decimation of BigBrainSym surface
addpath(genpath(bbwDir))
BB = SurfStatAvSurf({[bbwDir '/spaces/bigbrainsym/gray_left_327680_2009b_sym.obj'], ...
[bbwDir '/spaces/bigbrainsym/gray_right_327680_2009b_sym.obj']});
numFaces= 20484;
patchNormal = patch('Faces', BB.tri, 'Vertices', BB.coord.','Visible','off');
Sds = reducepatch(patchNormal,numFaces);
[~, bb_downsample] = intersect(patchNormal.Vertices,Sds.vertices,'rows');
BB10.tri = double(Sds.faces);
BB10.coord = Sds.vertices';
Rather than doing away with the other ~310,000 vertices though, we assign each of the removed vertices to the nearest maintained vertex, determined by shortest path on the mesh (ties are solved by shortest Euclidean distance). In this manner, all 320,000 vertices belong to one of ~10,000 parcels.
% For each vertex on BB, find nearest neighbour on BB10, via mesh neighbours
nn_bb = zeros(1,length(BB.coord));
edg = SurfStatEdg(BB);
parfor ii = 1:length(BB.coord)
nei = unique(edg(sum(edg==ii,2)==1,:));
if isempty(nei) && ismember(ii,bb_downsample)
nn_bb(ii) = ii;
else
while sum(ismember(nei, bb_downsample))==0
nei = [unique(edg(sum(ismember(edg,nei),2)==1,:)); nei];
end
matched_vert = nei(ismember(nei, bb_downsample));
if length(matched_vert)>1 % choose the mesh neighbour that is closest in Euclidean space
n1 = length(matched_vert);
d = sqrt(sum((repmat(BB.coord(1:3,ii),1,n1) - BB.coord(:,matched_vert)).^2));
[~, idx] = min(d);
nn_bb(ii) = matched_vert(idx);
else
nn_bb(ii) = matched_vert;
end
end
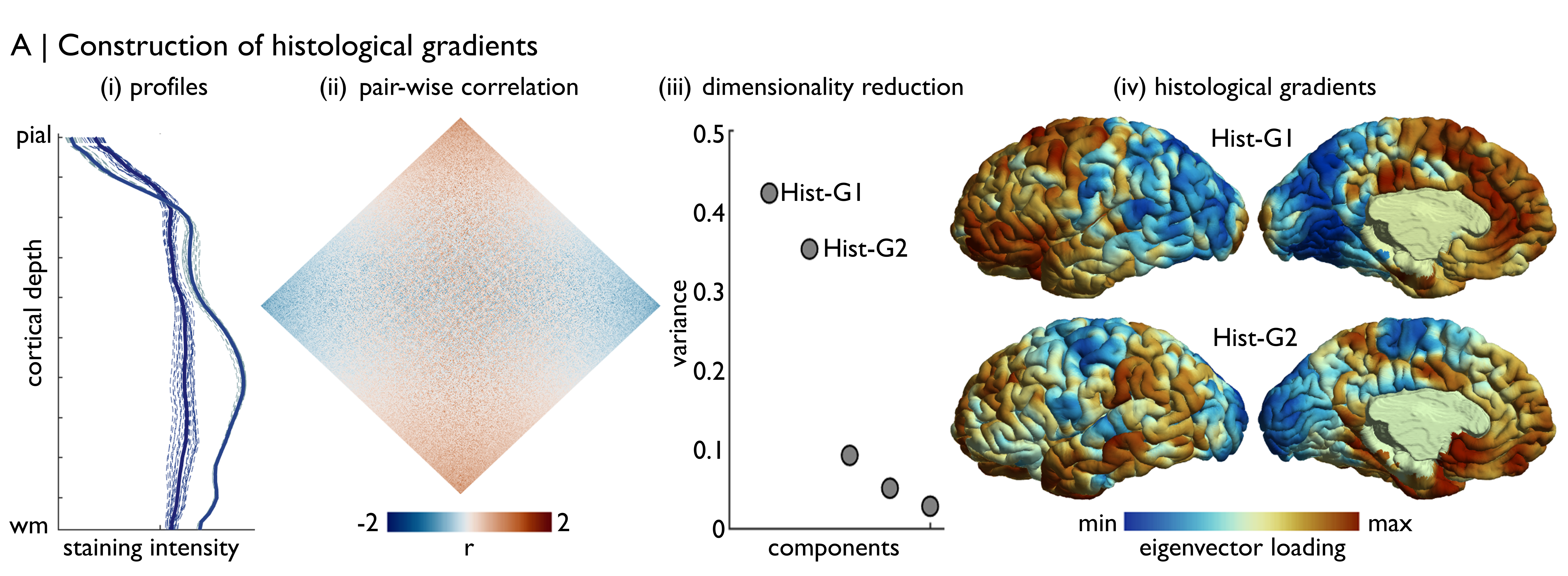
With the downsampling organised, we can move onto the construction of gradients. For BigBrain, we average staining intensity profiles within each parcel, then calculate the similarity between each pair of patch-average microstructure profiles, producing a microstructure profile covariance (MPC) matrix that reflects patch-wise cytoarchitectural similarity. Next, we calculate the normalised angle similarity between each row of the MPC matrix, which depicts patch-wise similarities in the patterns of MPC across the cortex. Then, we subject the normalised angle matrix to diffusion map embedding. Diffusion map embedding produces eigenvectors, describing the principle axes of variance in the input matrix, and resolve cortical gradients. Each eigenvector is accompanied by an eigenvalue that approximates the variance explained by that eigenvector. Here, the first two eigenvectors explain approximately 42% and 35% of variance, respectively. Projected on to the downsampled BigBrainSym surface, the first two eigenvectors illustrate an anterior-posterior and a sensory-fugal gradient. You may note that the eigenvector decomposition is different to our previous publication, where the sensory-fugal gradient explained more variance than the anterior-posterior gradient. In our previous work, we regressed the midsurface y-coordinate from the microstructure profiles because we observed a strong increase in intensity values with the y-coordinate (r = −0.68) that appeared related to the coronal slicing of BigBrain. In BigBrainWarp, we have opted for a more a conservative approach to data cleaning to avoid obscuring potential non-artefactual anterior-posterior variations in staining intensity.
% BigBrain profiles
MP = reshape(dlmread([bbwDir '/spaces/bigbrain/profiles.txt']),[], 50)';
% load downsampling indexing
load([bbwDir '/scripts/nn_surface_indexing.mat'], 'nn_bb');
% create MPC and gradient, using functions from the micaopen github
MPC = build_mpc(MP,nn_bb);
normangle = connectivity2normangle(MPC, 0);
[eigenvectors, results] = mica_diffusionEmbedding(normangle, 'ncomponents', 10);
eigenvalues = results.lambdas/sum(results.lambdas);
for ii = 1:2
Gmpc = BoSurfStatMakeParcelData(eigenvectors(:,ii), S, nn_bb); # re-expands to whole cortex using a SurfStat function (https://github.com/MICA-MNI/micaopen/)
lhOut = [bbwDir '/spaces/bigbrain/Hist-G' num2str(ii) '_lh.txt'];
rhOut = [bbwDir 'spaces/bigbrain/Hist-G' num2str(ii) '_rh.txt'];
writematrix(Gmpc(1:end/2)', lhOut)
writematrix(Gmpc((end/2)+1:end)', rhOut)
end
Next, we transform the BigBrain-derived gradients to fsaverage using BigBrainWarp
# run from within BigBrainWarp github repo
for ii in 1 2 : do
bigbrainwarp --in_space bigbrain --out_space fsaverage --wd /local/directory/for/output/ \
--in_lh spaces/bigbrain/Hist_G${i}_lh.txt \
--in_rh spaces/bigbrain/Hist_G${i}_rh.txt \
--desc Hist_G${i} \
--interp linear
done
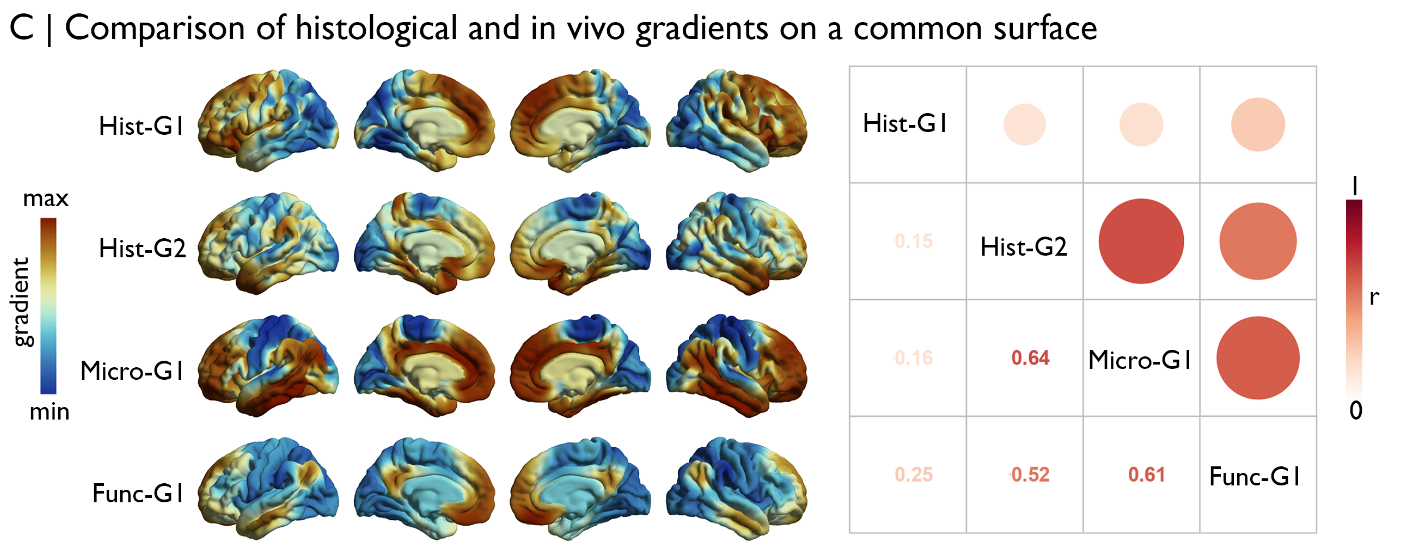
Et voila! The BigBrain-derived gradients are aligned to the MRI-derived gradients from the MICs data and can be statistically evaluated. The construction of the MRI-derived gradients is discussed at length in the micapipe documentation. Suffice to say, qT1 and rs-fMRI data from 50 healthy adults were registered to fsaverage, then parcellated using a downsampled 10k mesh (using the same procedure as shown above for BigBrain). The microstructural gradient was generated using the MPC approach, with quantitative T1 images rather than BigBrain, and the functional gradient was created from resting state functional connectivity (a la Margulies et al.,).
For example, we may assess spatial correspondence of the gradients using Spearman correlations. The statistical analysis should take into account the degree of downsampling, as such we subject the transformed histological gradients to the same parcellation as imaging-derived gradients. We find that there is strong correlation between the sensory-fugal BigBrain-derived gradient with the MRI-derived microstructural gradient (r=0.64), and both of these are moderately correlated with the functional gradient (both r>0.5). In contrast, the anterior-posterior BigBrain-derived gradient does not overlap with the MRI-derived gradients (all r<0.3).